Running a Spark Job Using PySpark on AWS EMR
What is Spark?
Spark is considered as a preferred data processing engine, primarily for usage in a vast range of situations. Data scientists and application developers integrate Spark into their own implementations in order to transform, analyze and query data at a larger scale. Functions which are mostly related with Spark, contain collective queries over huge data sets, machine learning problems and processing of streaming data from various sources.
What is PySpark?
PySpark is considered as the interface which provides access to Spark using the Python programming language. PySpark is basically a Python API for Spark.
What is EMR?
Amazon Elastic MapReduce, commonly known as EMR, is an Amazon Web Services mechanism for big data analysis and processing. This is established based on Apache Hadoop, which is a Java based programming framework which assists in the processing of huge data sets in a distributed computing environment. EMR also manages a vast group of big data use cases, such as bioinformatics, scientific simulation, machine learning and data transformations.
The Use Case
I’ve been mingling around with PySpark for the last few days and I was able to build a simple Spark application and execute it, as a step, in an AWS EMR cluster. The following functionalities were covered within this use case (See Figure 1).
Flowchart of the Above Functionalities
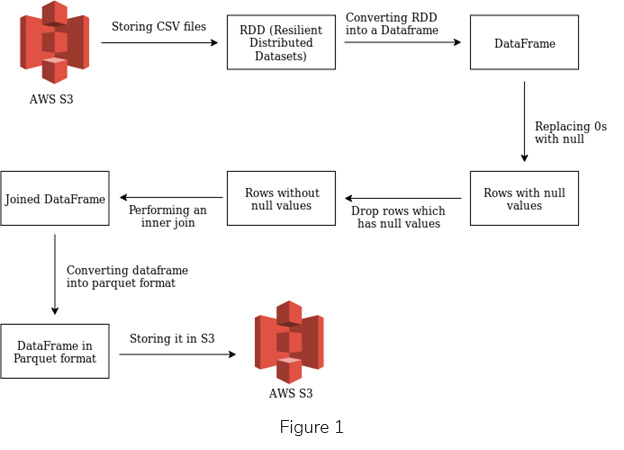
Let me explain each of the above functionalities by providing appropriate snippets.
1.0 Reading CSV files from AWS S3
This is where two files from an S3 bucket are being retrieved and will be stored in two data frames individually.
#importing necessary libraries
from pyspark.sql.functions import *
from pyspark.sql.types import StringType
from pyspark import SQLContext
from itertools import islice
from pyspark.sql.functions import col
#creating the context
sqlContext = SQLContext(sc)
#reading the first CSV file and store it in an RDD
rdd1= sc.textFile(“s3n://pyspark-test-kula/test.csv”).map(lambda line: line.split(“,”))
#removing the first row as it contains the header
rdd1 = rdd1.mapPartitionsWithIndex(
lambda idx, it: islice(it, 1, None) if idx == 0 else it
)
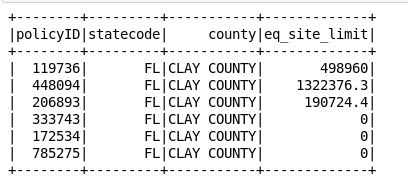
2.0 Converting an RDD (Resilient Distributed Datasets) into a data frame
#converting the RDD into a data frame
df1 = rdd1.toDF([‘policyID’,’statecode’,’county’,’eq_site_limit’])
#print the data frame
df1.show()
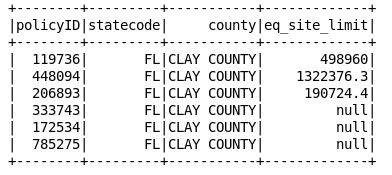
3.0 Replacing 0s with null values
#data frame which holds rows after replacing the 0s into null
targetDf = df1.withColumn(“eq_site_limit”, \
when(df1[“eq_site_limit”] == 0, ‘null’).otherwise(df1[“eq_site_limit”]))
targetDf.show()
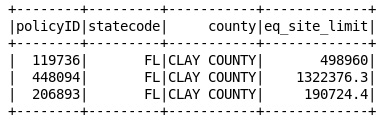
4.0 Dropping the rows which have null values
df1WithoutNullVal = targetDf.filter(targetDf.eq_site_limit != ‘null’)
df1WithoutNullVal.show()
Creating the second data frame
rdd2 = sc.textFile(“s3n://pyspark-test-kula/test2.csv”).map(lambda line: line.split(“,”))
rdd2 = rdd2.mapPartitionsWithIndex(
lambda idx, it: islice(it, 1, None) if idx == 0 else it
)
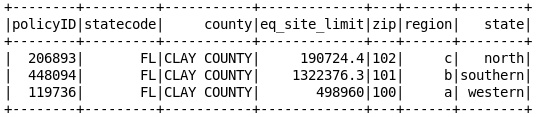
df2 = df2.toDF([‘policyID’,’zip’,’region’,’state’])
df2.show()
5.0 Performing an inner join on both data frames
innerjoineddf = df1WithoutNullVal.alias(‘a’).join(df2.alias(‘b’),col(‘b.policyID’) == col(‘a.policyID’)).select([col(‘a.’+xx)
for xx in a.columns] + [col(‘b.zip’),col(‘b.region’), col(‘b.state’)])
innerjoineddf.show()
6.0 Saving the joined data frame in the Parquet format back to S3
innerjoineddf.write.parquet(“s3n://pyspark-transformed-kula/test.parquet”)
Once we’re done with the above steps, we’ve successfully created the working Python script which retrieves two CSV files, stores them in different data frames and then merges both of them into one, based on some common column.
7.0 Executing the script in an EMR cluster as a step via CLI
We can now submit this Spark job in an EMR cluster as a step. To do this the following steps need to be followed:
1. Create an
EMR cluster
, which includes Spark, in the appropriate region
2. Once the cluster is in the
WAITING state, add the Python script as a step
3. Then execute this command from your CLI (Ref from the
doc)
aws emr add-steps — cluster-id j-3H6EATEWWRWS — steps Type=spark,Name=ParquetConversion,Args=[ — deploy-mode,cluster, — master,yarn, — conf,spark.yarn.submit.waitAppCompletion=true,s3a://test/script/pyspark.py],ActionOnFailure=
CONTINUE
If the above script has been executed successfully, it should begin the step in the EMR cluster, which you have mentioned. Normally it takes few minutes to produce a result, whether it’s a success or failure. If it’s a failure, you can probably debug the logs and see where you’re going wrong. Otherwise you’ve achieved your end goal.
Complete source-code: www.gist.github.com
References: www.docs.aws.amazon.com, www.ibmbigdatahub.com

Kulasangar Gowrisangar
Senior Software EngineerDeveloping a Custom Audit Trail and a Notification Service for a Workflow Based Serverless Application on AWS
READ ARTICLE
Human Emotions Recognition through Facial Expressions and Sentiment Analysis for Emotionally Aware Deep Learning Models
READ ARTICLE
